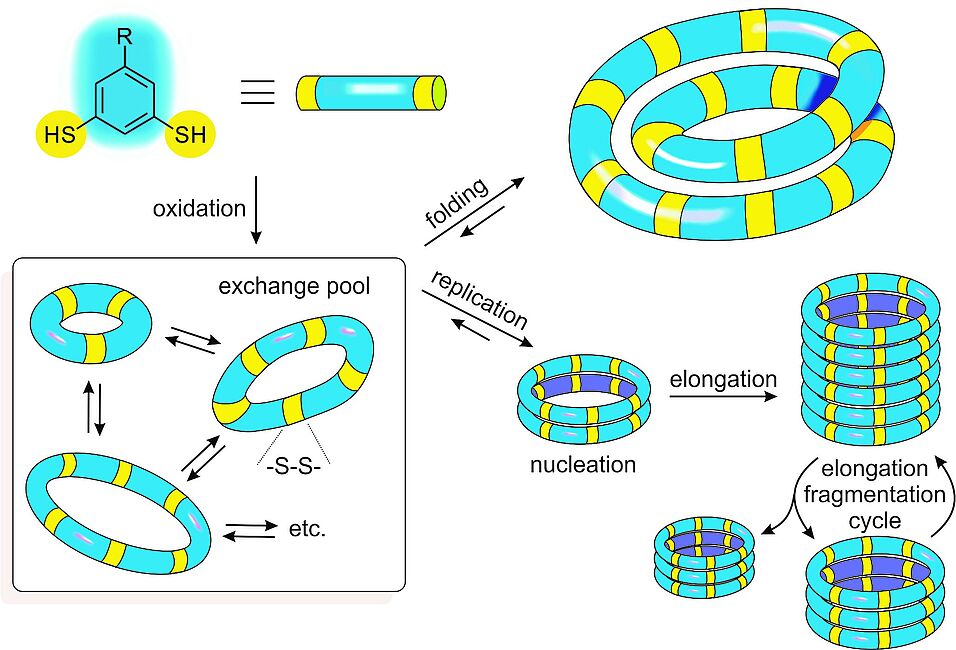
How the immense complexity of living organisms has arisen is one of the most intriguing questions in contemporary science. We have started to explore experimentally how organization and function can emerge from complex molecular networks in aqueous solution.1 We focus on networks of molecules that can interconvert, to give mixtures that can change their composition in response to external or internal stimuli. Noncovalent interactions within molecules in such mixtures can lead to the formation of foldamers.2,3 Molecular recognition between molecules in such mixtures leads to their mutual stabilization, which drives the synthesis of more of the privileged structures (Figure 1). As the assembly process drives the synthesis of the very molecules that assemble, the resulting materials can be considered to be self-synthesizing. Intriguingly, in this process the assembling molecules are replicating themselves, where replication is driven by self-recognition of these molecules in the dynamic network.4 The selection rules that dictate which (if any) replicator will emerge from such networks are starting to become clear.5 We have also witnessed spontaneous differentiation (a process akin to speciation as it occurs in biology) in a system made from a mixture of two building blocks.6 When such systems are operated under far-from-equilibrium flow conditions, adaptation of the replicators to a changing environment can occur.
Replicators that are able to catalyse reactions other than their own formation have also been obtained, representing a first step towards metabolism.7,8 Thus, the prospect of Darwinian evolution of purely synthetic molecules is tantalizingly close and the prospect of synthesizing life de-novo is becoming increasingly realistic.9
1. Li, J.; Nowak, P.; Otto, S. J. Am. Chem. Soc. 2013, 135, 25, 9222-9239.
2. Liu, B.; Pappas, C. G.; Zangrando, E.; Demitri, N.; Chmielewski, P. J.; Otto, S. J. Am. Chem. Soc. 2019, 141, 1685-1689.
3. Pappas, C. G.; Mandal, P. K. Liu, B.; Kauffmann, B.; Miao,X.; Komáromy, D.; Hoffmann, W.; Manz, C.; Chang, R.; Liu, K.; Pagel, K.; Huc, I.; Otto, S. Nature Chem. 2020, 12, 1180–1186.
4. Carnall, J. M. A.; Waudby, C. A.; Belenguer, A. M.; Stuart, M. C. A.; Peyralans, J. J.-P.; Otto, S. Science 2010, 327, 1502-1506.
5. Malakoutikhah, M.; Peyralans, J.J-P.; Colomb-Delsuc, M.; Fanlo-Virgos, H.; Stuart, M. C. A.; Otto, S.
J. Am. Chem. Soc. 2013, 135, 49, 18406-18417.
6. Sadownik, J. W.; Mattia, E.; Nowak, P.; Otto, S. Nature Chem. 2016, 8, 264–269.
7. Monreal Santiago, G.; Liu, K.; Browne, W. R.; Otto, S. Nat. Chem. 2020, 12, 603-607.
8. Ottelé, J.; Hussain, A. S.; Mayer, C.; Otto, S. Nat. Catal. 2020, 3, 547-553.
9. Adamski, P.; Eleveld, M.; Sood, A,; Kun, A.; Szilágyi, A.; Czárán, T.; Szathmáry, E.; Otto, S. Nat. Rev. Chem. 2020, 4, 386–403.
Prof. Sijbren Otto, Centre for Systems Chemistry, Stratingh Institute, University of Groningen, the Netherlands: s.otto@rug.nl, http://www.otto-lab.com/
Join us via ZOOM: https://univienna.zoom.us/j/92576672420?pwd=ZkIvMm1RbUM0aUtGV3VQemRFZXlnUT09
Meeting-ID: 925 7667 2420
Code: 891211
Via SIP: 92576672420@zoomcrc.com